Introduction to dental plaque
Dental plaque is a complex community of micro-organisms that forms on the surfaces of teeth and restorations and has been implicated as the primary etiological factor in the development of periodontal diseases 1, 2. So far, more than 700 different bacterial species have been identified from the human oral cavity, and a majority of them are associated with dental plaque 3-5. Presently, there is a strong evidence in favor of association of the plaque with the development of inflammatory periodontal diseases viz. gingivitis and periodontitis 6-10. Furthermore, it has been shown that periodontal diseases are the result of complex interactions between the host and the bacteria and the prevalence of certain bacteria (considered as periodontal pathogens) increases at the sites where active periodontal destruction is taking place 8. Research has demonstrated that plaque develops as a well-organized structure on the tooth surface in the form of a biofilm. Biofilms have been described in many systems since Van Leeuwenhoek examined the “animalcules” in the plaque on his own teeth in the seventeenth century, but the general theory of biofilm predominance was not promulgated until 1978 when Costerton invented the word “biofilm”, referring to the matrix-enclosed bacterial community 11. The slime layer that forms in the dental unit waterlines is an example of biofilm familiar to most dental professionals. Biofilm can also be found lining oil pipelines, fish tanks, indwelling catheters, internal implants, contact lenses, and prosthetic devices. Biofilm is composed of microcolonies of bacterial cells (15-20% by volume) that are non-randomly distributed in a shaped matrix or glycocalyx (75-80% volume). In the following discussion, we shall discuss various aspects of dental plaque.
Definition of dental plaque
Dental plaque is defined as a diverse community of micro-organisms found on the tooth surface as a biofilm, embedded in an extracellular matrix of polymers of host and microbial origin (Marsh PD, 2004) 12.
Dental plaque can also be defined as a specific but highly variable structural entity resulting from sequential colonization and growth of microorganisms of various strains and species on the surfaces of teeth, restorations and other parts of the oral cavity, embedded in the extracellular matrix composed of bacterial metabolic products and substances from serum, saliva, and blood.
Classification of dental plaque
In relation to the gingival margin:
Supra-gingival
Sub-gingival
Attachment to the tooth surface:
Attached
Un-attached
In relation to health and disease:
Health-associated
Disease-associated
Development of dental plaque
The development of dental plaque was studied by Löe et al. (1965) 13 in an experimental gingivitis study on dental students who were restrained from all oral hygiene measures for a period of three weeks. Following observations were made in the study,
- Soon after the tooth surface is thoroughly cleaned, it rapidly gets covered with a glycoprotein deposit referred to as “pellicle” which is derived from salivary constituents like albumin, lysozyme, amylase, immunoglobulin A, proline-rich proteins, and mucins, which are selectively adsorbed onto the tooth surface. This is the first step in plaque formation.
- The tooth surface coated with the dental pellicle is now colonized by bacteria referred to as “primary colonizers”. These include Gram-positive bacteria such as Streptococcus mitis, Streptococcus sanguis, Streptococcus oralis, Streptococcus mutans and Actinomyces viscosus 14-16. It must be noted that during the initial colonization, although Actinomyces species are the most prevalent among Gram-positive rods, but these constitute less than 10% of the total viable counts. A lag phase or relatively slow growth phase during initial plaque formation has been reported 14, 16, 17. The reason suggested for this lag phase is the relatively slow growth rate of the Actinomyces species. It is important to note here that during bacterial aggregation, interactions between the bacterial cell wall and surfaces (including other cell walls) are primarily influenced by interfacial electrostatic (e.g., repulsion, attraction) and van der Waals forces 18, 19. The initial colonizers can be isolated from cleaned tooth surfaces within 60 minutes after cleaning the teeth. The attachment of these organisms to pellicle is facilitated by……. Contents available in the book……… Contents available in the book…….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…

- After the initial colonization, the plaque increases in size by two distinct mechanisms: 1) the multiplication of bacteria already attached to the tooth surface, and 2) the subsequent attachment and multiplication of new bacterial species to the cells of bacteria already present in the plaque mass. With plaque maturation, the microflora of plaque becomes more diverse. The proportions of Actinomyces species and other Gram-positive bacilli increases, which provide receptors to other bacterial species that were previously unable to colonize on the pellicle coated tooth surface. The metabolic products of pioneer communities attached to the dental pellicle alter the local environment and make the conditions suitable for the growth of fastidious bacterial species. Gradually, the conditions become more suitable for the growth of obligate anaerobes. The new bacteria that attach to plaque are referred to as “secondary colonizers”. These secondary colonizers primarily include Gram-negative species such as Fusobacterium nucleatum, Prevotella intermedia, and Capnocytophaga species. A key property of these microorganisms appears to be the ability to adhere to Gram-positive species already present in the existing plaque mass. These organisms can be found in dental plaque typically after 1-3 days of its accumulation. The composition of bacterial microflora in plaque changes over time due to a series of complex interactions and this process has been termed as ‘microbial succession’.
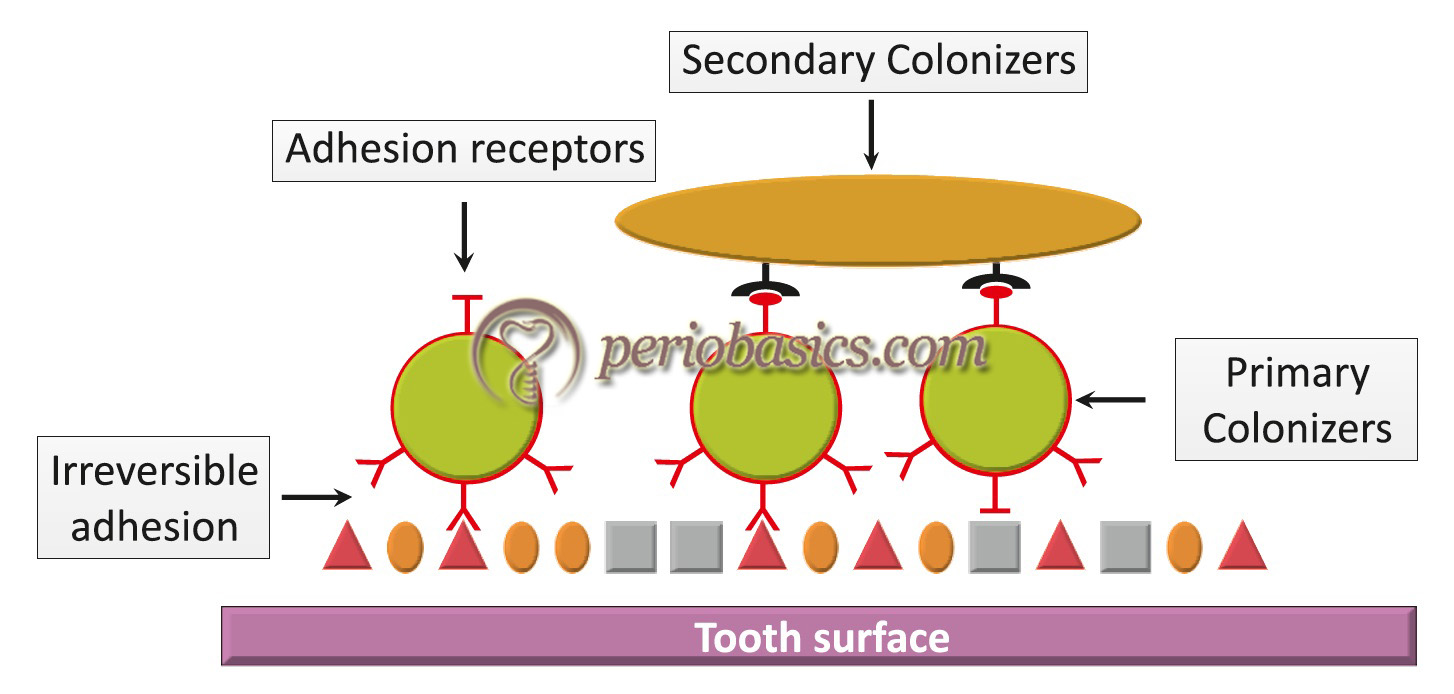
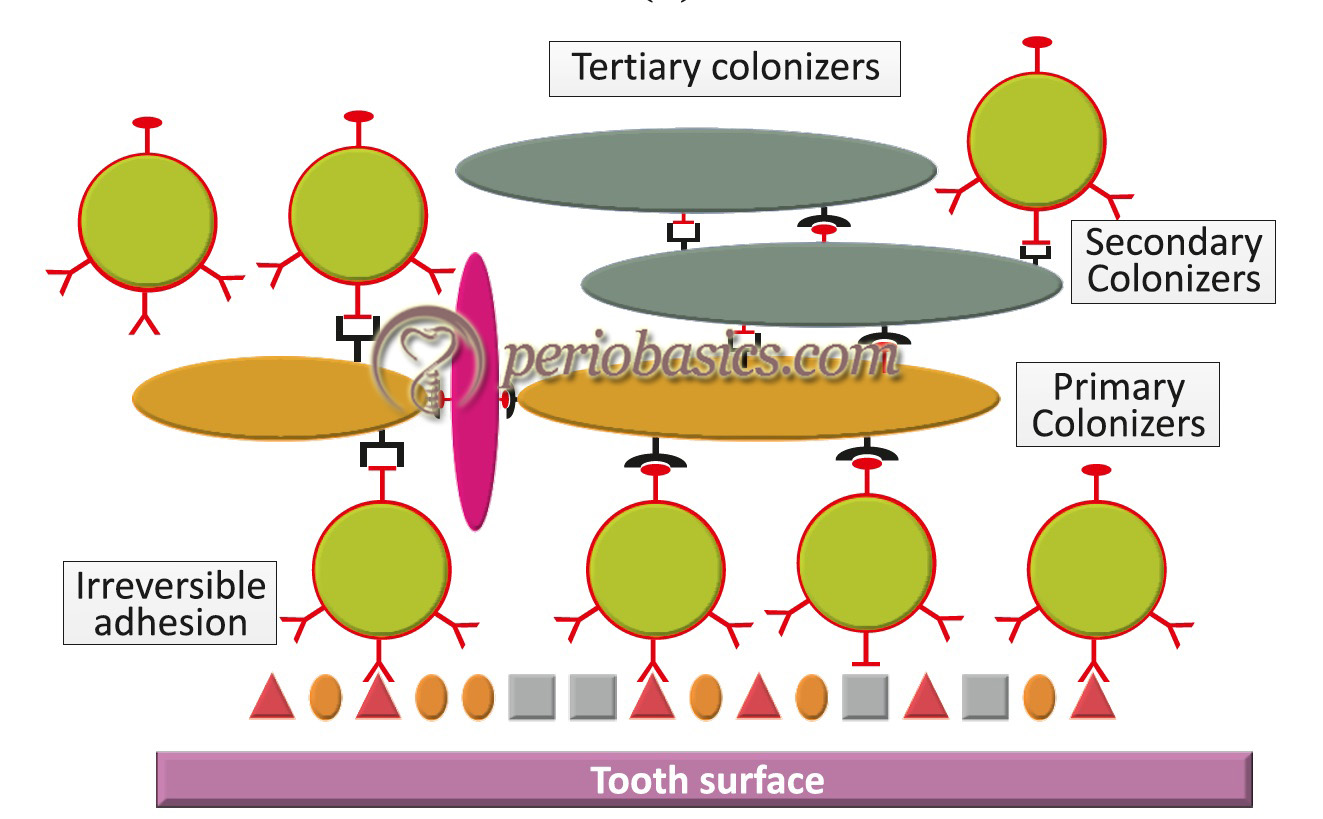
- Further maturation of dental plaque leads to colonization of primarily Gram-negative species. These colonizers are referred to as “tertiary colonizers” which include Porphyromonas gingivalis, Campylobacter rectus, Eikenella corrodens, Aggregatibacter actinomycetemcomitans, and the oral spirochetes (Treponema species). At this point of time, the structure of plaque is typically complex with complex patterns of bacterial cells of cocci, rods, fusiform, filaments, and spirochetes. A “corn-cob” like arrangement of microorganisms can be observed microscopically. Recently, a “hedgehog” like arrangement ……. Contents available in the book……… Contents available in the book…….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
It must be remembered that bacterial colonization on any surface is significantly affected by various surface properties which can be broadly divided into macroscopic and microscopic surface properties. Let us discuss these properties in detail.

Role of macroscopic surface characteristics during plaque formation
Various macroscopic surface properties that affect bacterial colonization include surface energy, zeta potential, and hydrophobicity. All these properties are interrelated to each other.
Surface energy and surface tension
Surface energy is defined as the sum of all the intermolecular forces that are present on the surface of a material. It is the sum of the degree of attraction or repulsion forces of a material surface which it exerts on other materials. More is the surface energy; more is the tendency to attract. Another important term that must be understood is surface tension. Surface tension can be defined as the resistance of a fluid to deform or break. The lower is the surface tension, lesser is the resistance to deform or rupture. When the substrate has high surface energy (high tendency to attract) and the adhesive has a low surface tension, a good wettability is ensured. These are important properties affecting initial colonization of bacteria on the tooth surface 21-23.
Surface energy can be calculated by measuring the in vitro contact angle measurements of the substratum 21, 22 or of the substratum and the microorganisms 23. Surfaces that are very hydrophobic or very hydrophilic tend to retain few microorganisms. The adsorption of microorganisms on a surface depends on both the critical surface tension of the substratum and the flow rate of saliva 22. Surfaces with intermediate surface tension (35-38 mN/m) have been shown to accumulate the highest number of microorganisms from whole saliva 14-16. It must be noted that when a microorganism attaches to a surface, the overall surface energy of the system changes. Adhesion is favored, if it is accompanied by a decrease in the free surface energy of the whole system 24. Furthermore, microorganisms with high free surface energy tend to adhere to surfaces with high surface free energy and vice versa. It has been demonstrated that …….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…
Another important factor determining the plaque formation is surface roughness. It has been shown that the influence of surface roughness is more important in plaque formation in vivo than surface free-energy per se. In an in vitro study, it has been demonstrated that rough surface (13-15 μm) was more heavily colonized as compared to a smooth surface (0.75 μm). Furthermore, plaque on a rougher surface was at a more mature stage after six days as compared to that on a smoother surface 27. It has also been demonstrated that root surfaces are more heavily colonized by microorganisms as compared to enamel surface 14, 17.
Zeta potential
Zeta potential is defined as the potential difference existing between the surface of a solid particle immersed in a conducting liquid (e.g. water) and the bulk of the liquid. Zeta potential of any microorganism is determined by the nature and the number of the ionogenic groups on the cell surface and ionic strength and pH of the surrounding medium. Bacterial adhesion is facilitated by a low negative charge on the bacterial surface. Various initial colonizers during plaque formation, such as strains of S. mutans, S. sanguis, S. salivarius, Actinomyces viscosus, and Actinomyces odontolyticus show relatively high surface free energies (range, 99-128 mJm-2) and carry a negative surface charge which facilitates their adsorption over enamel surface 28.
Hydrophobicity
Hydrophobicity of various microorganisms varies consider-ably. However, it is an important property as far as microbial adherence is concerned. Hydrophobic organisms are attracted towards the solid surfaces due to rejection from the aqueous phase. There are several techniques which can be used to determine the degree of hydrophobicity of the bacterial cells. One commonly used technique is BATH (bacterial adherence to hydrocarbons) method, proposed by Rosenberg (1984) 29, which is now more generally known as MATH (microbial adherence to hydrocarbons). Hydrophobicity favors bacterial adhesion. It has been shown that the hydrophobic Streptococci strains adhere better to hydroxyapatite surfaces in vitro as compared to the less hydrophobic strains 30. However, it does not mean that more the hydrophobicity of the bacterial cell surface; more is its adherence. In a study, it was found that both hydrophobic and hydrophilic strains of Streptococcus mutans were implanted equally well on the enamel surface 31. There-fore, hydrophobicity cannot be considered as a definitive property for bacterial adhesion.
Role of microscopic surface characteristics during plaque formation
The microscopic surface characteristics that affect bacterial adhesion to the tooth surface include surface characteristics of salivary components and microbial adhesins.
Salivary components
Salivary components can be considered as a major factor during initial bacterial adhesion to the tooth surface. Salivary oligosaccharide-containing glycoproteins have been shown to provide receptors for oral Streptococci in the salivary pellicle 32. Furthermore, salivary proline-rich protein 1 and statherin have been shown to provide receptor sites for type 1 fimbriae of Actinomyces viscosus 33. However, on the contrary, salivary components which favor bacterial adhesion as a component of dental pellicle, inhibit bacterial adhesion when they are present in saliva solution by binding to microbial adhesins 34, 35. This phenomenon can be explained on the basis of certain ‘cryptitopes’ or previously hidden molecular segments which may serve as receptors once the molecule gets adsorbed on a surface. In other words, we can say that conformational changes occur in a molecule when it gets adsorbed on a surface, exposing certain hidden receptors which provide attachment to the bacteria. These ‘cryptitopes’ may also be revealed due to enzymatic cleavage of terminal residues of the molecule.
Microbial adhesins
The adhesion of a microorganism on a surface is directly related to its inherent capability to adhere to a surface 36. The majority of the microorganisms possess a large number of adhesins on their surface which may bind stereochemically to the complementary receptors on the interacting surface. These adhesins are often associated with surface appendages like fimbria, pilli or high-molecular-weight proteins extending from the microbial surface. It has been proved by electron microscopic studies that the early colonizers are attached to the tooth surface by microbial surface fimbriae 17. In vitro studies have suggested that the attachment of S. mutans to saliva-coated hydroxyapatite surface is mediated by its fibrillar, hydrophobic surface-protein P1 37, 38. However, when P1-deficient S. mutans strains were evaluated for adherence as compared to P1-intact strains on rat teeth, both were found to adhere on enamel surface 38. These findings suggest the diversity of adhesive mechanisms of bacterial attachment on a surface.
Physiochemical process of bacterial adhesion
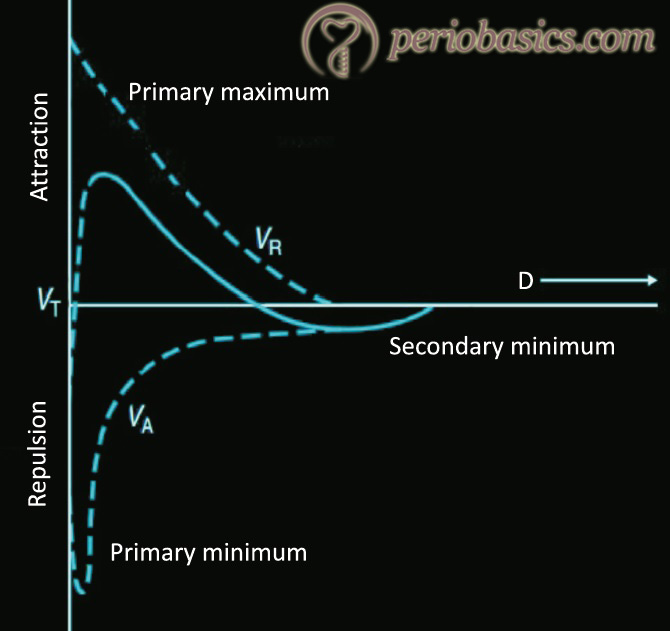
The physiochemical interactions between the bacterial cell and the surface of the substratum have been explained by thermodynamic model and DLVO (Derjaguin, Landau, Ver-wey, Overbeek) theory. The thermodynamic theory explains the bacterial adhesion on the basis of interfacial free energies of the interacting surfaces (bacterial cell surface and substratum surface). It does not take into consideration the role of electrostatic interaction 39. On the other hand, DLVO theory describes the interaction energies between the interacting surfaces, based on electrostatic interaction and van der Waals forces and their decay with separation distance 40. According to this theory, the total interactive energy …….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
Interspecies interactions and metabolic diversity of microorganisms in dental plaque
As already stated, dental plaque consists of various strains and species of microorganisms, thus making a complex living community. There are various types of interactions that occur in between these organisms. Various animal and in vitro studies have revealed a multitude of bacterial interspecies interactions, which can be of following types 41-46,
- Competitive interactions between bacteria for nutrients.
- Synergistic interactions, which may stimulate the growth or survival of one or more residents.
- Antagonistic interaction, where one resident inhibits the growth of another.
- Neutralization interaction, where virulence factors produced by one microorganism get neutralized by another resident.
- Interference in the growth-dependent signaling mechanisms of one organism by another.
The microorganisms within dental plaque can be categorized as saccharolytic or asaccharolytic. Saccharolytic microorganisms in the supragingival sites ferment carbo-hydrates primarily into lactic acid and create an acidic environment temporarily. Conversely, in the subgingival sites, asaccharolytic microorganisms metabolize nitrogenous compounds derived from gingival crevicular fluid (GCF) and create a neutral pH and anaerobic environment abundant in short-chain fatty acids and ammonia. The metabolism of amino acids by asaccharolytic microorganisms leads to the production of sulfur compounds, the major components responsible for oral malodor. These metabolic changes lead to alterations in the environmental conditions and introduce more pathogenic microorganisms into the microbial plaque.
Supragingival plaque is dominated by Streptococcus and Actinomyces species. With the help of adhesions and receptors, they adhere to the saliva-coated tooth surface and use salivary components as nutrients 47. These bacteria are saccharolytic and degrade the carbohydrates derived from foods through the Embden-Meyerhof-Parnas (EMP) pathway to form lactic, formic, acetic, succinic and other organic acids, and concomitantly consume oxygen by NADH oxidase community. This leads to the acidification of the environment and anaerobic conditions. In due course of time, supragingival pH can reach around 4.
Subgingival plaque is dominated by asaccharolytic and anaerobic and/or proteolytic bacteria, such as Fusobacterium, Eubacterium, Campylobacter, Prevotella, and Porphyromonas. This is because, as the gingival crevice deepens the environment becomes more anaerobic with neutral pH. Continuous efflux of gingival crevicular fluid (GCF), nutritionally rich in nitrogenous compounds such as amino acids, peptides and proteins provide nutrition to the microorganisms. Proteolytic bacteria like P. gingivalis (releasing gingipains) and P. intermedia degrade nitrogenous compounds into small peptides and amino acids by cell membrane-bound and/or extracellularly secreted proteases. P. gingivalis prefers neutral pH 48, 49 while P. intermedia and F. nucleatum are capable of growth at acidic and neutral pH 50 and are frequently found in supragingival plaque. P. gingivalis and Prevotella intermedia use acetic-succinic pathway for the production of acetic and succinic acids from aspartic acid. P. gingivalis also uses propionic-butyric pathway responsible for the production of propionic and butyric acids from glutamic acid.
Oral malodor is mainly produced by volatile sulfur compounds (VSC) produced through oral bacterial metabolism of sulfur amino acids. Major components of VSC are hydrogen sulfide (H2S), methyl mercaptan and dimethyl sulfide. Periodontal disease-related bacteria including Porphyromonas and Prevotella are responsible for VSC production in malodor subjects with periodontal diseases while commensal bacteria such as Actinomyces and Veillonella are responsible for H2S production in malodor subjects without periodontal disease 51, 52.
Present research indicates that the metabolic products by plaque bacteria may promote or inhibit the growth of other species 53-55. For example, interspecies synergistic relation-ships that can be observed within a plaque biofilm are as follows,
- Protoheme produced by Campylobacter rectus functions as a growth factor for P. gingivalis.
- Formate produced by Prevotella melaninogenica stimulates the growth of C. rectus.
- Lactic acid produced by Streptococcus and Actinomyces is needed for the metabolism of Veillonella which, in turn, produce menadione that favors the growth of Porphyromonas and Prevotella.
- Fatty acids produced by Fusobacterium are needed for the growth and metabolism of Treponema 56.
The metabolic interdependency of two or more bacterial species has been termed as “metabolic mutualism”. It has been observed that mutually beneficial bacterial species show better growth as co-culture as compared to mono-species culture. One of the well-established examples of this relationship is …….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…
Antagonism of one bacterial species by the other is also seen in plaque biofilm. One bacterial species may prevent the colonization of other species by using receptors, which may be needed for the attachment of other bacterial species 59. Further-more, one bacterial species can produce substances that may inhibit the growth of other bacterial species or may inhibit the expression of their virulence factors. For example, hydrogen peroxide produced by S. sanguinis inhibits the growth of A. actinomycetemcomitans while A. actinomycetemcomitans produces a bacteriocin that inhibits S. sanguinis 60. Hence, in a microbial community such as plaque biofilm, both synergistic as well as antogonistic interactions occur between bacterial species.
The behavior of planktonic bacteria is different as compared to the same bacteria in the biofilm. In a study done by Sanui et al. (2009) 61, it was observed that, when present in a biofilm, S. mutans expressed more fructan hydrolase, exo-beta-D-fructosidase, fructanase, and cell division protein FtsG, as compared to its planktonic state. On the other hand, the expression of antigen I/II, glucosamine-fructose-6-phosphate aminotransferase I and chaperonin GroEL was found to be reduced in the biofilm. The metabolic requirements of bacteria are also different in biofilm as compared to its planktonic state. For example, it has been observed that growth of S. mutans in planktonic state is enhanced when grown in the presence of low concentration of nicotine (0.25-0.5 mg/ml) and is inhibited at high concentration (2-4 mg/ml), whereas, when in a biofilm, high concentration of nicotine enhances the growth of S. mutans 62.
It should be noted here that certain bacterial species which are not in complete biological compatibility with each other develop a physiological state of mutual compatibility. For example, lactate produced by S. gordonii is used by A. actinomycetemcomitans as an alternative source of carbon 63. On the other hand, S. gordonii produces H2O2 which is cytotoxic to A. actinomycetemcomitans. In response to H2O2, A. actinomycetemcomitans demonstrates a …… Contents available in the book…….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…
Changing views of plaque
The concept of dental plaque and its etiological potential in periodontal diseases has changed a lot because of our increasing knowledge about etiopathogenesis of periodontal diseases. The following paragraphs explain how the specific plaque hypothesis was the first one to be accepted, then non-specific plaque hypothesis was explained and finally specific plaque hypotheses was re-accepted 65.
Specific plaque hypothesis
The period from 1880 to 1930 is known as the ‘golden age of microbiology’ 66. During this period, the pathogens that caused many systemic infections of medical importance were identified. Researchers also looked for a single, specific cause of oral diseases. Assuming plaque contained the micro-organism that caused periodontal disease, dental scientists studied plaque in search of the causative agent. Towards the end of the 19th century…… Contents available in the book…….. Contents available in the book…….. Contents available in the book…….. Contents available in the book…
Non-specific plaque hypothesis
The 1930’s ushered in a different view of the role of plaque and its microorganisms in the etiology of periodontal disease. Dental scientists believed that periodontal disease was linked with some constitutional defect in the individual 67. Studies conducted between 1930 and 1970 failed to identify any specific organism as the etiologic agent of periodontal diseases. These negative findings formed a basic tenet of the non-specific hypothesis which suggested that periodontal disease is due to subgingival accumulation of micro-organisms beyond a threshold that can be limited by mechanisms of host resistance 68. This theory has subsequently evolved into the specific plaque hypothesis, which postulates that certain bacteria are the etiologic agents of distinct forms of periodontal diseases (explained later) 69. Mechanical irritants such as calculus and overhanging restorations were also thought to play a major role in the pathogenesis of periodontal disease. The belief that a single microbial agent caused periodontal disease was replaced by nonspecific plaque theory. Non-specific plaque hypothesis held that the entire bacterial flora in the plaque played a role in periodontal destruction rather than any specific bacteria. All plaque was viewed as bad plaque. Furthermore, more plaque meant more disease. Plaque control was viewed as an essential measure to limit the production of gingival irritants that lead to inflammation and periodontal destruction 70. Identification of specific microorganisms was not important. Stringent plaque control was important, and it became the focus of periodontal therapy.
Return of specific plaque hypothesis
The 1960’s marked a return to specific plaque hypothesis. Researchers were successful in showing that periodontal disease could be transmitted between hamsters 71. The electron microscope confirmed that spirochetes were present in the connective and epithelial tissues of patients with acute necrotizing ulcerative gingivitis in contrast to healthy controls 72. Believing there were differences in plaque brought about by different species, scientists again returned to the search for a specific microbial periodontal pathogen and treatment aimed at the causative agent 67.
Both non-specific and later specific plaque hypothesis were described by Walter Loesche 69 in the year 1976.
Ecological plaque hypothesis
This hypothesis has been proposed by Marsh (1994) 45. To understand ecological plaque hypothesis, it is essential to understand the development of plaque as a biofilm. Let us first discuss the concept of a biofilm.
Development of plaque as biofilm
The formation and establishment of a biofilm on a surface is governed by a variety of physical, chemical and biological processes. As already discussed, various macroscopic and microscopic surface properties play a vital role in the whole process of biofilm formation. There is a specific and ordered sequence of events that results in the formation of a structurally and functionally well-organized biofilm structure 2. Distinct stages in the dental biofilm formation are,
- Dental pellicle formation where macromolecules present within the bulk of oral fluid settle onto a substrate through gravitational force or movement of flow. Weak electrostatic forces also play a significant role in this process.
- Reversible adhesion involving weak, long-range physicochemical interactions between the bacterial cell surface and the pellicle, which can lead to a stronger adhesin-receptor mediated attachment. Founder microbes attach to the tooth surface primarily through weak van der Waals forces. The reversible adhesion is affected by various factors including pH, temperature, surface energy and electrostatic forces. It has been observed that microbial adhesion strongly depends on the hydrophobic-hydrophilic properties of the interacting surfaces 73.
- Irreversible adhesion involves the physical adhesion of bacteria on the tooth surface by overcoming the physical repulsive forces. The bacteria attach irreversibly to the tooth surface using fimbriae or pili that are present on their outer surface. The microorganisms excrete a mixture of exo-polysaccharides and DNA, called the extra-cellular matrix (ECM) which aids in their attachment to the surface and gives them protection from the surrounding environmental conditions.

- Colonization involves an increase in the bacterial population, which results in the formation of a “proto-biofilm” that grows in size by two ways: through the founder microorganisms dividing as well as other microorganisms joining the biofilm from the surrounding environment. The microorganisms that join the biofilm aren’t necessarily the same species of bacteria as the founder microorganisms. They might not even be bacteria and can be other microorganisms like fungi and protozoa. Some of the microbes that join biofilms do not produce exopolysaccharides or secrete DNA and might not have the fimbriae that allow them to bind to founder organisms. These organisms join the biofilm by attaching to the sticky extracellular matrix (ECM) of the biofilm.
- Co-adhesion involves attachment of secondary colonizers to the already attached cells through cell surface interactions 74. It must be understood that bacterial co-adhesion is very specific. Bacterial species attach specifically to other bacterial species. Some bacterial species can attach to many other bacterial species, while others may attach only to a few of them. For example, F. nucleatum can attach to various bacterial species, thus acting as a bridge or nucleus during plaque formation.
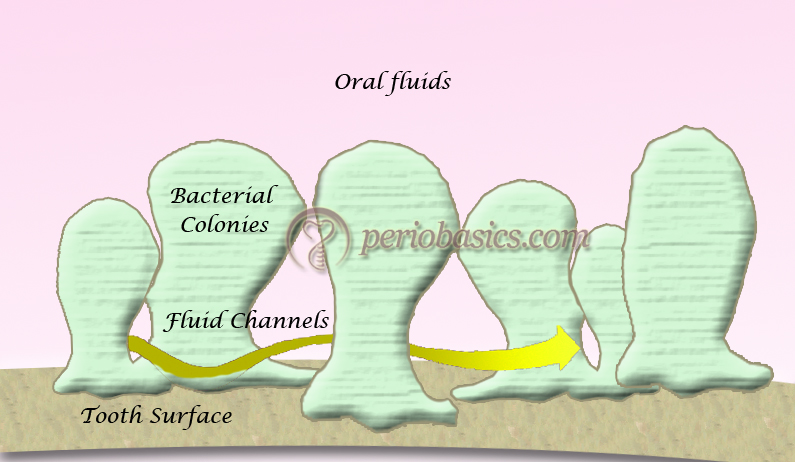
- Multiplication and biofilm maturation results in a well-established biofilm. A mature biofilm has a rough, irregular surface that contains many individual colonies of non-uniform, mushroom-shaped or finger-like columns surrounded by fluid-filled channels in which nutrients, enzymes, and waste products circulate 75. The water channels permit the passage of nutrients and other agents throughout the biofilm acting as a primitive ”circulatory” system. A mature biofilm becomes .. Contents available in the book…….. Contents available in the book…….. Contents available in the book…….. Contents available in the book….
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
It can be summarized from the above discussion that the life cycle of a biofilm is characterized by the attachment of planktonic bacteria to a surface or by migration or division of sessile cells to cover an empty region of the surface, production of extracellular polymeric substances to adhere cells irreversibly to the substrate, and then by additional extracellular polymeric substances production, cellular motility and reproduction, and phenotypic differentiation to produce a mature, thick and spatially structured biofilm 78. The bacteria in biofilms are phenotypically distinct from their genomically-identical planktonic counterparts. Bacteria in biofilms can be up to 1000 times more resistant to antibiotics, and less conspicuous to the immune system because antigens are hidden and key ligands are suppressed 79.
Basic biofilm properties
- Co-operating community of various types of micro-organisms.
- Microorganisms are arranged in micro-colonies.
- Micro-colonies are surrounded by a protective matrix.
- Within the micro-colonies are differing environments.
- Microorganisms have a primitive communication system.
- Microorganisms in a biofilm are resistant to antibiotics, antimicrobials, and host response.
Quorum sensing
The term ‘quorum-sensing’ was first used in a review by Fuqua et al. (1994) 81, which essentially reflected the minimum threshold level of individual cell mass required to initiate a concerted population response. The signal molecule that is used for communication was called as ‘autoinducer’, owing to its origin inside the bacterial cell. The desired response can be arrived at by attainment of quorum employing the autoinducer and the process was labeled as ‘autoinduction’. Quorum sensing is the regulation of bacterial gene expression in response to fluctuations in local signal concentration. This is done via the production and release of molecular autoinducers that increase in concentration as a function of cell density in a liquid culture; when quorum-sensing cells detect a minimum threshold stimulatory concentration of an autoinducer, gene expression changes 82, 83.
Quorum sensing in bacteria “involves the regulation of the expression of specific genes through the accumulation of signaling compounds that mediate intercellular communication” 84. Quorum sensing is dependent upon cell density. With few cells, signaling compounds may be produced at low levels; however, autoinduction leads to increased concentration as cell density increases. Once the signaling compounds reach a threshold level (quorum cell density), gene expression is activated.
Signaling is not the only way of transferring information in biofilms. The high density of bacterial cells growing in biofilms facilitates the exchange of genetic information between the cells of the same species and across the species or even genera. For example, genes for antibiotic resistance can be transferred through this efficient process. Other ways of communication between bacteria in a biofilm are,
- Through small diffusible peptides.
- Through horizontal gene transfer.
- Through autoinducer 2, which mediates the communication between Gram-positive and Gram-negative bacteria.
Autoinducer 2 (AI-2) plays a very important role in multispecies aggregation in a biofilm by acting as a universal intergeneric signaling molecule 85. The lux S gene which expresses AI-2 is conserved among many bacterial species including S. mutans, Streptococcus gordonii, Streptococcus oralis, P. gingivalis, A. actinomycetemcomitans and other oral microorganisms 86, 87. It was demonstrated in one study that when grown as monoculture biofilm in …… Contents available in the book…….. Contents available in the book…….. Contents available in the book….
Mechanisms of increased antibiotic resistance of organisms in a biofilm
As already stated, it has been recognized for considerable periods of time that microorganisms growing in the biofilms are more resistant to antibiotics than the same species growing in a planktonic (unattached) state 102-106. Traditionally, minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) terms have been used to describe the minimum concentration of an antimicrobial agent which is able to stop the growth and kill the microorganisms grown in a culture media, respectively. Given, that bacterial resistance to antimicrobials increases many times in a biofilm, the terms biofilm inhibitory concentration (BIC) and biofilm killing concentration (BKC) have been proposed to more accurately predict bacterial killing in a biofilm. However, these terminologies have not yet been widely accepted.
Although the mechanisms of resistance of micro-organisms to antibiotics growing in biofilms are not entirely clear 107, following mechanisms have been proposed to explain the antibiotic resistance in biofilm microorganisms:
- One important mechanism of resistance appears to be the slower rate of growth of bacterial species in the biofilms, which makes them less susceptible to many but not all antibiotics 108-111. The matrix performs a ”homeostatic function”, such that cells deep in the biofilm experience different conditions such as hydrogen ion concentration or redox potentials than the cells at the periphery of the biofilm or cells growing planktonically. The growth rates of these deeper cells will be decreased, allowing them to survive better than faster-growing cells at the periphery when exposed to antimicrobial agents 1. Slower- growing bacteria often overexpress ”nonspecific defense mechanisms” including shock proteins and multi-drug efflux pumps (arcAB) and demonstrate increased exopolymer synthesis 112.
- The exopolymer matrix of a biofilm, although not a significant barrier by itself to the diffusion of antibiotics, does have certain properties that can retard diffusion.
Extracellular enzymes such as β-lactamases, formaldehyde lyase, and formaldehyde dehydrogenase may become trapped and concentrated in the extracellular matrix, thus inactivating susceptible, typically positively charged, hydrophilic antibiotics. Some antibiotics such as the macrolides, which are positively charged but hydrophobic in nature, are unaffected by this process. - Hydrodynamics 113 and the turnover rate of the micro-colonies also affect antibiotic effectiveness 114.
- Recently, the notion of a subpopulation of cells within a biofilm that are ”super-resistant” was proposed. Such cells could explain remarkably elevated levels of resistance to certain antibiotics that have been suggested in the literature. Brooun et al. (2000) 109 examined the contribution of multi-drug resistance pumps to antibiotic resistance of microorganisms grown in biofilms. These ”pumps” can extrude chemically unrelated antimicrobial agents from the cell.
With this basic description about biofilm formation and its properties, let us now discuss the ecological plaque hypothesis,
The ecological plaque hypothesis
This hypothesis combines the key elements of specific and non-specific plaque hypothesis. As already stated, according to specific plaque hypothesis there are certain micro-organisms responsible for the progression of a diseases and according to non-specific plaque hypothesis, it is a heterogeneous mixture of microorganisms that plays a crucial role in the disease progression. The key elements of “ecological plaque hypothesis” are 115:
- The selection of “pathogenic” bacteria is directly coupled to the changes in the environment.
- Diseases need not have a specific etiology; any species with relevant traits can contribute to the disease process.
According to this hypothesis, changes in the environmental conditions lead to an ecological shift. This ecological shift provides favorable conditions for the growth of pathogenic microorganisms or expression of pathogenic traits. The evidence for this hypothesis comes from the mixed cultures studies 116, 117 and from other work which provides an argument that plaque-mediated diseases are a consequence of imbalances in the resident microflora resulting from an enrichment within the microbial community of these “oral pathogens”. Bacterial species such as Actinomyces play an important role in maintaining stability and balance in the microflora community. These species produce acid in the oral cavity by fermenting carbohydrates, thus altering the pH of the oral cavity. However, the homeostatic system of the oral cavity is usually able to regulate the change in pH, thereby maintaining a pH close to 7 118.
If the change in the pH of oral cavity persists for a longer duration of time, the balance between plaque and host immune response is disturbed. Oral conditions like increased availability of fermentable carbohydrates or decreased salivary flow favors acid production by microorganisms, thus resulting in the shift in oral flora from predominantly Gram-positive bacteria to predominantly Gram-positive facultatively anaerobic microflora followed by Gram-negative obligate anaerobic or micro-aerophilic flora. Moreover, any bacterial species can be pathogenic since the change in the environment may facilitate expression of virulence factors associated with that species 119, 120. In an experiment, Marsh (1991) 121 demonstrated that carbohydrate challenge at neutral pH had little effect on bacterial colonization but when pH was allowed to fall as a result of carbohydrate metabolism, number of bacterial species including S. mutans, Lactobacillus, and Veillonella species increased and became the dominant species.
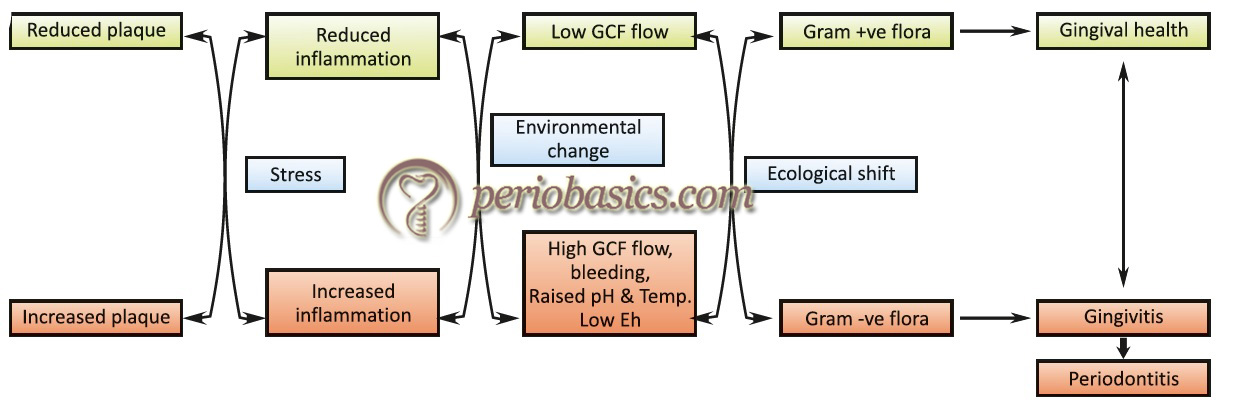
So, according to ecological plaque hypothesis disease can be prevented not only by targeting the putative pathogens directly, e.g. by antimicrobial or anti-adhesive strategies, but also by interfering with the selection pressure responsible for their enrichment 76.
Keystone pathogen hypothesis
This hypothesis is based on the fact that certain micro-organisms have the ability to influence their surrounding environment which is disproportionate to their relative overall abundance 122-124. This hypothesis has been proposed by Hajishengallis et al. (2012) 125 to explain periodontopathogenic plaque in the oral cavity. Authors have proposed that even at low-abundance certain microbial pathogens can change the composition of the bacterial plaque and cause inflammatory periodontal disease. For example, it has been shown that P. gingivalis can manipulate the host immune response which facilitates not only its own survival but also the survival of the entire microbial community 125, 126. These observations were made by studying the “red complex” as described by Socransky et al. (1998) 8. It was observed in mouse models that even with very low presence, P. gingivalis (<0.01% of the total bacterial count in the plaque) was able to alter the plaque composition thereby facilitating the development of periodontitis. Furthermore, it was observed that in germ-free mice, P. gingivalis was able to colonize but was not able to induce periodontitis without the presence of other bacterial species indicating P. gingivalis alone cannot induce periodontitis 127. The finding that P. gingivalis acts as keystone pathogen has also been supported by studies on rabbit models 128 and non-human primates 129.
When the disease progresses, the relative numbers of these keystone pathogens increases which further facilitates disease progression 8. Furthermore, it has been demonstrated that even when the absolute number of these keystone pathogens increases, their overall presence in plaque reduces when compared with the total bacterial load which increases as plaque accumulates in periodontitis 130.
The host immune response is also an important compo-nent of keystone plaque hypothesis. There are various chemical mediators and cell surface receptors that play a key role during initiation of the inflammatory response (details available in “Host-microbial interactions in periodontal diseases”). Expression of IL-8 by epithelial cells is responsible for …… Contents available in the book…….. Contents available in the book…….. Contents available in the book….
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
- Toll-like receptor (TLR) response manipulation,
- Interleukin 8 (IL-8) subversion, and
- Interference with complement system.
In vitro, the TLR response is manipulated by P. gingivalis by its two different lipopolysaccharide lipid A structures viz. Type I and Type II, where, Type I is a TLR4 agonist which activates the immune system, while Type II is a TLR4 antagonist which inhibits the immune response against P. gingivalis 144, 145. P. gingivalis secretes a serine phosphatase that inhibits the synthesis of IL-8 146. This process has been termed as “local chemokine paralysis” and has been explained as the mechanism by which P. gingivalis hampers the recruitment of PMN’s thus preventing formation of an adequate neutrophil wall 147. T. denticola, another microorganism in the red complex is also able to manipulate IL-8 response but the mechanism has not been well understood 148.
The third and the most well-understood mechanism of immune response manipulation by keystone pathogens is interference with the complement system. This interference is executed by P. gingivalis through its membrane-bound and soluble arginine-specific cysteine proteinases referred to as “gingipains” (discussed in “Microbiology of periodontal diseases”).The complement system plays an important role in recognizing and destroying microorganisms 149. Evasion of innate immune response is an important property of a microorganism which makes it capable of colonization and proliferation in the host. P. gingivalis has been shown to cleave complement factors C3 and C5 into active fragments C5a (cell activator) and C3b (phagocytosis enhancer). Gingipains degrade these fragments, causing loss of their function 150.
Although most of the evidence for keystone plaque hypothesis has been derived from the research on P. gingivalis, most of the other presently known or unknown keystone organisms may also manipulate the innate immune response by the above-stated mechanisms or other mechanisms which are still not known to us.
Dental plaque associated with periodontal health
Dental plaque is established on the tooth surface soon after cleaning of the teeth. In periodontal health, a balance is established between microorganisms in plaque and host defense mechanism. Plaque-associated with periodontal health also helps the host by occupying the niches which would otherwise be harbored by periodontal pathogens. Bacterial genera belonging to Streptococcus and Actinomyces colonize on tooth surface soon after cleaning the teeth which explain their prevalence in dentitions which are well maintained 151. The prevalence of Gram-negative facultative anaerobes is more in areas with active periodontal disease. In darkfield and phase contrast microscopic investigations it has been demonstrated that in healthy periodontal sites, the prevalence of species belonging to spirochetes and motile rods is 1-3% and 1-6%, respectively and for coccoid bacterial species, it is 62-79% 152-155. Furthermore, after the treatment of periodontitis, the relative counts of Gram-negative facultative microorganisms have been shown to reduce with a concomitant increase in cocci 154-157.
Dental plaque associated with periodontal diseases
As already stated, most of the periodontal pathogens are Gram-negative facultative microorganisms. P. gingivalis, F. nucleatum, T. forsythia, P. intermedia, spirochetes and species belonging to A. actinomycetemcomitans have been shown to be associated with periodontal disease sites. Socransky et al. (1998) 8 have described various microbial complexes associated with periodontal status in health and disease. A detailed description of these complexes is available in “Microbiology of periodontal diseases”.
Plaque around dental implants
The microflora around successful implants is similar to healthy sulci, while that associated with failing implants is similar to periodontally diseased sites. Various studies suggest that anaerobic plaque bacteria (associated with periodontitis) may have an adverse effect on peri-implant tissue health 158. The implant sulcus can be colonized by bacteria from saliva or neighboring periodontal pockets.
The microbiota associated with healthy peri-implant tissues closely resembles that of the flora associated with gingival health. Experimental studies have shown that plaque accumulation, gingival indices, and probing depths increased around the implants in the same way as they do around the normal tooth when restrained from regular oral hygiene procedures 159. Hence, the accumulation of plaque around implants can lead to peri-implantitis. Microbiological studies have demonstrated that successful implants are colonized by Gram-positive cocci, whereas failing implants are colonized by a large number of Gram-negative anaerobes 160, 161. Aggregatibacter actinomycetemcomitans, Prevotella intermedia, Porphyromonas gingivalis, Fusobacterium species, Campylobacter rectus have all been associated with failing implants 162-164.
A healthy peri-implant socket is mainly colonized by oral Streptococci which constitute from 45% to 86% of supra- and subgingival peri-implant sulcus microbiota. Actinomyces naeslundii, Actinomyces oris and Actinomyces meyeri, as well as Neisseria and Rothia species, are also frequently isolated 165. A shift from healthy peri-implant sulcus to a diseased peri-implant pocket is associated with the increased presence of cocci, motile bacilli and spirochetes. Peri-implant mucositis and peri-implantitis sites have been shown to demonstrate an increase in the proportion of periodontopathogenic bacteria, mostly from the orange complex: F. nucleatum, P. intermedia and Eubacterium species 166, 167.
Conclusion
The biophysiology of microorganisms in a biofilm is very complex. An autogenic succession occurs in microorganisms with the maturation of plaque which involves complex interactions that take place in between microorganisms of various strains and species. With the maturation of plaque, various periodontal pathogens increase in number and start producing virulence factors and thus initiate periodontal inflammation. From the above discussion it should be clear that dental plaque should never be considered as a static ecosystem but a dynamic ecosystem which is affected by its surrounding environment. Plaque is considered as the primary etiological factor for the development of periodontal diseases and its control is the first step in the treatment of periodontal diseases. Because plaque is a complex community of micro-organisms, there is a lot of scope for research to enlighten its behavior in periodontal health and disease.
References
References available in the hard-copy of the website
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.